Dates : 2016 Nov. 24th, 10:00 -- Nov. 25th 16:00
Place : Lille University, Cité scientifique, Villeneuve d'Ascq.
Organization :
BioComputing group of
the computer science lab CRIStAL of the
university of Lille
(Cedric Lhoussaine, Joachim
Niehren and Cristian
Versari)
Topic of the workshop:
Given that metabolism is a central aspect of all living systems, the domain of metabolomics has become a large research domain in molecular biology. In the last 10 years, this has raised increasing interest in the formal modeling and analysis of metabolic networks. A typical example is flux balance analysis, but also the control of the metabolic fluxes has become subject to formal modeling and prediction approaches. The objective of the workshop is therefore to bring together researchers from computer science, biology, chemistry, and mathematics, who are working on different aspects of the modeling of metabolic networks.
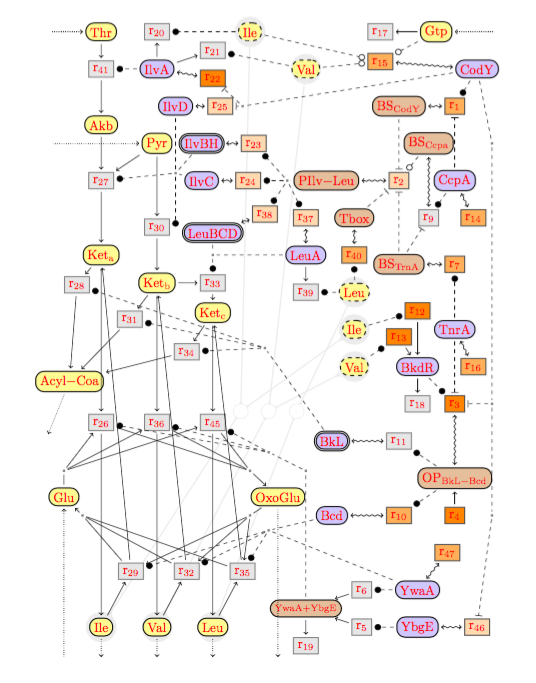
A model of the BCAA metabolic network of B.~Subtilis
Bioss will support the workshop fees and the hotel costs. Only the travel costs are left to the participants. If you are interested to join the workshop, please contact the organizers by Email to bio-computing-metabolism@lists.gforge.inria.fr
Program
The precise schedule is yet to be announced but we will be starting at 10:00 on Nov. 24th and finish at 16:00 on Nov. 25th.
- Frank Brueggeman TBA
- Olivier Bernard Dynamic metabolic modelling of microalgae. Also presents work of Caroline Baroukh.
- François Coutte Surfactine overproduction and how the models for metabolism of B.Subtilis. Joint work with Cristian Versari, Joachim Niehren, Philippe Jacques, and Debarun Dali.
- Daniel Kahn Discussing metabolic interpretation of metagenomic data.
- Joachim Niehren Structural simplification of reaction networks in partial steady states. Joint work with Guillaume Madelaine, Cédric Lhoussaine, and Elisa Tonello.
- Jean-Pierre Mazat Small is beautiful. Human scale metabolic model(s).
- Nathalie Poupin Exploring metabolic modulations using genome scale network modelling and omics data.
- Sabine Peres and Philippe Dague SAT-based approaches for metabolic pathway analysis.
- Anne Siegel Reconstruction d'un réseau métabolique à partir de données brutes.
- Moritz von Stosch An elementary mode constrained latent variable method for reaction flux data analysis.
- Rajeev Khoodeeram A formal model of metabolism to decipher Crabtree effect. Joint work with Jean-Yves Trosset and Gilles Bernot.
- Emilie Allart Change prediction of reaction networks with partial kinetic information and the elementary modes. Joint work with Cristian Versari and Joachim Niehren.
Participants
Frank Brueggeman Vrije
Universitat Amsterdam
Caroline
Baroukh
Université de Toulouse
Olivier
Bernard
Inria Sophia-Antipolis
Philippe Dague Université Paris Sud
Daniel Kahn
INRA Lyon
Rajeev KHOODEERAM I3S, UNICE
Cédric Lhoussaine
Université de Lille
Jean-Pierre Mazat
Université Bordeaux
Joachim Niehren Inria
Lille
Nathalie Poupin
INRA Toulouse
Sabine Peres Université Paris Sud
Elisabeth Remy Université de
Marseille
Anne Siegel Inria Rennes
Moritz von
Stosch
Newcastle University
Jean-Yves
Trosset
Sup’Biotech, Paris Sud
Cristian Versari
Université de Lille\
Dernière modification le 24/11/2016